The standard optimisation algorithms described above are written
in terms of basic commands, which may be put to much more use in
allowing the user to write his own Tcl script, which may contain
things like if-statements and loops.
The most flexible way to use the package is thus to invoke the
component commands directly from a Tcl script. To allow recursive or
reentrant optimisation algorithms to be constructed, an
object-oriented design has been adopted. An optimiser object is
created using the optimiser command, the name of the
object is then a valid Tcl command and methods are invoked by the syntax
| Method |
Args |
Function |
| px |
|
print current parameter position |
| sp |
|
test point as suitable restart point-tester is the
RMS of the gradient |
| ss |
|
save the start structure (only for the zopt
function, named zmatrix.start) |
| sr |
|
save the best available structure (only for the zopt function, named zmatrix.best) |
| wr |
name |
write object to file, attaching label. Valid labels are start, best, vis |
| f |
|
compute function value |
| g |
|
compute gradient |
| fdg |
|
compute gradient by finite-difference of energy |
| eg |
|
compute gradient without frozen hessian eigenmode
contributions |
| fg |
|
compute function and gradient |
| gn |
|
get current gradient norm (into
parm(gnorm)) |
| grms |
|
get current gradient rms (into no_parm(grms)) |
| tg |
|
test quality of gradient |
| ug |
|
transform gradient with hessian eigenvectors |
| h |
|
compute analytic hessian (for gamess only) |
| fdh |
|
generate hessian by finite differences of gradients
with a one-point formula for all variables |
| fdh2 |
|
generate hessian by finite differences of gradients
with a two-point formula for all variables |
| fd |
i |
generate hessian row and column for variable i by finite differences of gradients
with a one-point formula |
| fd2 |
i |
generate hessian row and column for variable i by finite differences of gradients
with a two-point formula |
| gh |
|
generate diagonal guess-hessian from simple rules |
| efdh |
i j ... |
modify hessian by finite-differencing along
i'th, j'th, k'th, etc. hessian eigenmodes and back-transform the hessian |
| efdh2 |
i j ... |
modify hessian by two-point finite-differencing along
i'th, j'th, k'th, etc. hessian eigenmodes and back-transform the hessian |
| sh |
m |
set hessian matrix from matrix m |
| shv |
i a |
set diagonal hessian value for variable i to a |
| mh |
|
condition hessian to have the correct inertia by
massageing the eigenvalues |
| sih |
m |
set inverse hessian from matrix m |
| sh1 |
|
set hessian matrix to be the unit matrix |
| sih1 |
|
set inverse hessian to be the unit matrix |
| dh |
|
diagonalise hessian |
| sm |
|
set (first usage) or select (all subsequent usages)
hessian eigenvector mode to be followed in TS search |
| sms |
|
set (first usage) or map (all subsequent usages)
hessian eigenvector modes unto starting modes |
| um |
|
update hessian eigenvalue mode to be followed in TS
search |
| hc |
|
determine the hessian inertia ( i.e. the number
of positive, negative, and zero eigenvalues) |
| ih |
|
invert hessian |
| iih |
|
invert the inverse hessian |
| dih |
|
diagonalise inverse hessian |
| pu |
|
Powell-update the hessian |
| bfu |
|
BFGS-update the hessian |
| bfui |
|
BFGS-update the inverse hessian using the GAMESS-UK
code |
| u0 |
|
BFGS estimate and first update of the inverse hessian |
| u |
|
BFGS-update the inverse hessian |
| msu |
|
Murtagh-Sargent-update the hessian |
| msui |
|
Murtagh-Sargent-update the inverse hessian |
| pmsu |
|
Hybrid Powell/Murtagh-Sargent-update the hessian |
| s |
|
take current step |
| ovls |
|
calculate overlap between current and previous step |
| rx |
|
reset the position vector to that of the previous
step |
| sl |
s |
set step length to s |
| cl |
s |
calculate step length and set it to s if it exceeds
s |
| clv |
s |
calculate search length and set it to s if it exceeds
s |
| rs |
s |
restrict step, such that largest element equals s |
| tr |
|
determine trust region by comparing actual and
predicted energy changes |
| ee |
|
estimate energy change from second-order model
(valid hessian assumed) |
| gs |
|
set step vector to steepest descent direction |
| es |
iv |
set step vector to inverse hessian eigenvector iv |
| evs |
iv |
set step vector to hessian eigenvector iv |
| pfm |
i |
project i'th hessian eigenvector (mode) out of the
step-vector |
| gda |
|
add current point to diis space |
| gdd |
i |
delete i'th (first point 0) point from diis space |
| gddg |
|
delete point with highest gradient rms from diis space |
| gdp |
|
print all current diis data points and gradients |
| gds |
|
evaluate diis step |
| gdw |
|
write diis space (points and gradients to files
newopt.gdxr and newopt.gdgr, respectively |
| gdr |
m1 m2 |
read diis space from matrices m1 (points) and
m2 (gradients) |
| cgr |
|
calculate conjugate gradient restart step vector |
| cg |
|
calculate conjugate gradient step vector
(Shanno-Phua code) |
| cgs |
|
calculate conjugate gradient step vector |
| n |
|
set step vector by newton method |
| prfo |
|
calculate (p)rfo step vector |
| ls |
|
line search step |
| ls2 |
|
line search step with at least two trial points |
| lsf |
|
line search step with function values only |
| lsg |
|
line search step with function and gradient values |
| tc |
|
check for convergence |
| d |
|
diagnose the current point for sensibility |
| sym |
m |
symmetrize square matrix m |
| lv |
|
list current variables and their gradients |
| pun |
name |
append the current structure to punch-file name for
viewing |
| praxis |
|
invoke PRAXIS optimizer |
| log |
|
print optimisation log (steps, line searches, updates) |
| x |
|
free memory |
The above commands operate on a group of Matrix data structures,
listed in the following table. In many cases it is not necessary to
access the data explicitly--since an entire optimisation can be
performed in terms of the operations given above. However, a series of
Tcl commands are available if one wants to
operate on the Matrix data structures directly.
NOTE: $opt.st, $opt.yt, and $opt.yi are defined only in a conjugate-gradient
optimisation.
The following table describes the elements of the Tcl array
no_parm that are used to control the operation of the primitive
commands. The first series of elements may be set during the
optimisation; the second series are generated by the various commands
and may be accessed to control the optimisation. The default values,
where given, are those applied in the control scripts, (as invoked by
the newopt and hessian commands. When writing custom
optimisation algorithms many will need to be set explicitly.
| Element |
Usage |
Default |
| function |
function to be invoked |
|
| args |
Tcl list of arguments to be passed to the function |
|
| method |
type of calculation |
finite_difference ( hessian); bfgs ( newopt) |
| threshold |
threshold-value for recalculation of hessian for
selected degrees of freedom |
|
| status |
Tcl return value |
0 |
| verbose |
output flag |
0 |
| del |
displacement in finite-difference formulae |
0.001 |
| maxstep |
maximum number of steps in search |
200 |
| conv_maxgrad |
maximum gradient convergence criterion |
0.001 |
| conv_rmsgrad |
RMS gradient convergence criterion |
0.0002 |
| conv_maxstep |
maximum parameter step convergence criterion |
0.001 |
| conv_rmsstep |
RMS parameter step convergence criterion |
0.0002 |
| toler_energy |
energy forced-convergence criterion |
1.0e-10 |
| list_variables |
full listing of variables after each step |
yes |
| step |
maximum step-length |
0.3 |
| lsopt |
type of line-search algorithm |
ls |
| ls_eps1 |
line-search convergence parameter 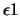 |
0.9 |
| ls_eps2 |
line-search convergence parameter 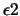 |
0.001 |
| trust_region |
vary maximum step length according to predictive
power of the second-order model |
inactive |
| stepmin |
minimum step length with trust-region method |
0.01 |
| stepmax |
maximum step length with trust-region method |
0.5 |
| follow_mode |
hessian eigenvalue mode to be followed
in TS search (use normal counting) |
0 ( i.e. minimisation) |
| update_mode |
redefine mode to be followed to be the step taken |
off (keep starting mode) |
| hessian_update |
define update method of the hessian |
powell |
| fin_diff_eigenmodes |
recalculate part of the hessian by
finite-differencing selected eigenmodes if inertia deteriorates |
no |
| keep_correct_hessian |
disregard update if hessian inertia
deteriorates |
no |
| recompute_hessian |
number of steps after which hessian is to be recomputed |
maxstep+1 ( i.e. no recalculation of the hessian) |
| recompute_method |
method by which to compute the hessian |
finite_difference |
| condition_hessian |
hessian conditioning |
off |
| run_diagnostics |
check sensibility of the current point |
off |
| old_energy |
energy of previous point |
|
| new_energy |
energy of current point |
|
| best_step |
index of step with lowest RMS gradient |
|
| trust_steps |
number of steps taken without RMS gradient getting
smaller |
10 |
| set_restart |
flag to write best restart structure after each step |
yes |
| thresh_prfo |
threshold for contribution of hessian eigenmodes to prfo step |
1.e-4 |
| conv_prfo |
convergence criterion for prfo shift parameter |
1.e-4 |
| contribute_prfo |
cutoff on hessian eigenvalue for contributing modes to prfo bracketing scheme |
1.e-4 |
| freeze_soft_modes |
flag for freezing near-zero eigenvalue hessian modes in Baker algorithm |
0 |
| soft_mode |
criterion on eigenvalue for considering a hessian eigenmode soft |
1.e-3 |
| freeze_modes |
flag for freezing selected hessian modes in Baker algorithm |
0 |
| mode_list |
Tcl list of indices of modes to freeze |
" " |
| min_diis_dimension |
minimum size of GDIIS space |
|
| max_diis_dimension |
maximum size of GDIIS space |
nvar/2 |
| read_diis_space |
flag for reading in GDIIS matrices |
0 |
| diis_positions |
name of matrix with GDIIS positions |
" " |
| diis_gradients |
name of matrix with GDIIS gradients |
" " |
| maxlength |
maximum allowed step length |
0.3 |
| nvar |
number of degrees of freedom in the optimisation
problem |
|
| hessian_ok |
indicator of correct hessian inertia |
no |
| best_hessian |
number of incorrect hessian eigenvalues |
|
| pos |
number of positive hessian eigenvalues |
nvar |
| gnorm |
gradient norm |
|
| grms |
gradient rms |
|
| map_modes |
Tcl list of map of current hessian eigenmodes on starting
modes |
|
![]()