The iron-molybdenum (FeMoco) and iron-vanadium cofactors (FeVco) in the molybdenum/vanadium nitrogenase enzymes catalyze the reduction of dinitrogen at ambient temperature and pressure to ammonia according to the reaction equation: N2 +8H+ 8e– +16MgATP→ H2 + 2NH3+16MgADP. How this complex 8-electron process is carried out by these enzymes is still a mystery.
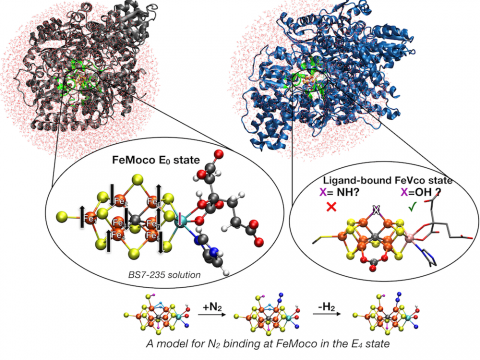
In a series of studies we have explored by QM/MM calculations, all performed with ChemShell, the resting state geometric and electronic structure of FeMoco in MoFe protein [1], the identify of the ligand in ligand-bound FeVco [2] and models for the N2-binding E4 state of FeMoco [3]. QM/MM methodology was shown to be critical for an accurate resting state FeMoco structure and to uncover the nature of ligand X in the structure of a recent ligand-bound iron-vanadium cofactor, FeVco in vanadium nitrogenase. Most recently, we have put forward a new model for the 4-electron reduced E4 state of FeMoco. Our model contains 2 hydrides that bridge Fe atoms no. 2 and 6 with a terminal sulfhydryl group and features favorable N2 binding and a subsequent reductive elimination step.
[1] B. Benediktsson and R. Bjornsson, “QM/MM Study of the Nitrogenase MoFe Protein Resting State: Broken-Symmetry States, Protonation States, and QM Region Convergence in the FeMoco Active Site” Inorg. Chem., 2017, 56, 13417-13429.
[2] B. Benediktsson, A. Th. Thorhallsson, and R. Bjornsson, “QM/MM calculations reveal a bridging hydroxo group in a vanadium nitrogenase crystal structure” Chem. Comm., 2018, 54, 7310-7313.
[3] A. Th. Thorhallsson, B. Benediktsson, R. Bjornsson, “A model for dinitrogen binding in the E4 state of nitrogenase” Chem. Sci., 2019, 10, 11110-11124.
